Cell confinement,cell confiner 细胞限制器装置-单细胞力学研究利器
型号:cell confiner
联系人:李胜亮
联系电话:18618101725
品牌:法国
细胞限制器-cell confiner(Cell confinement)
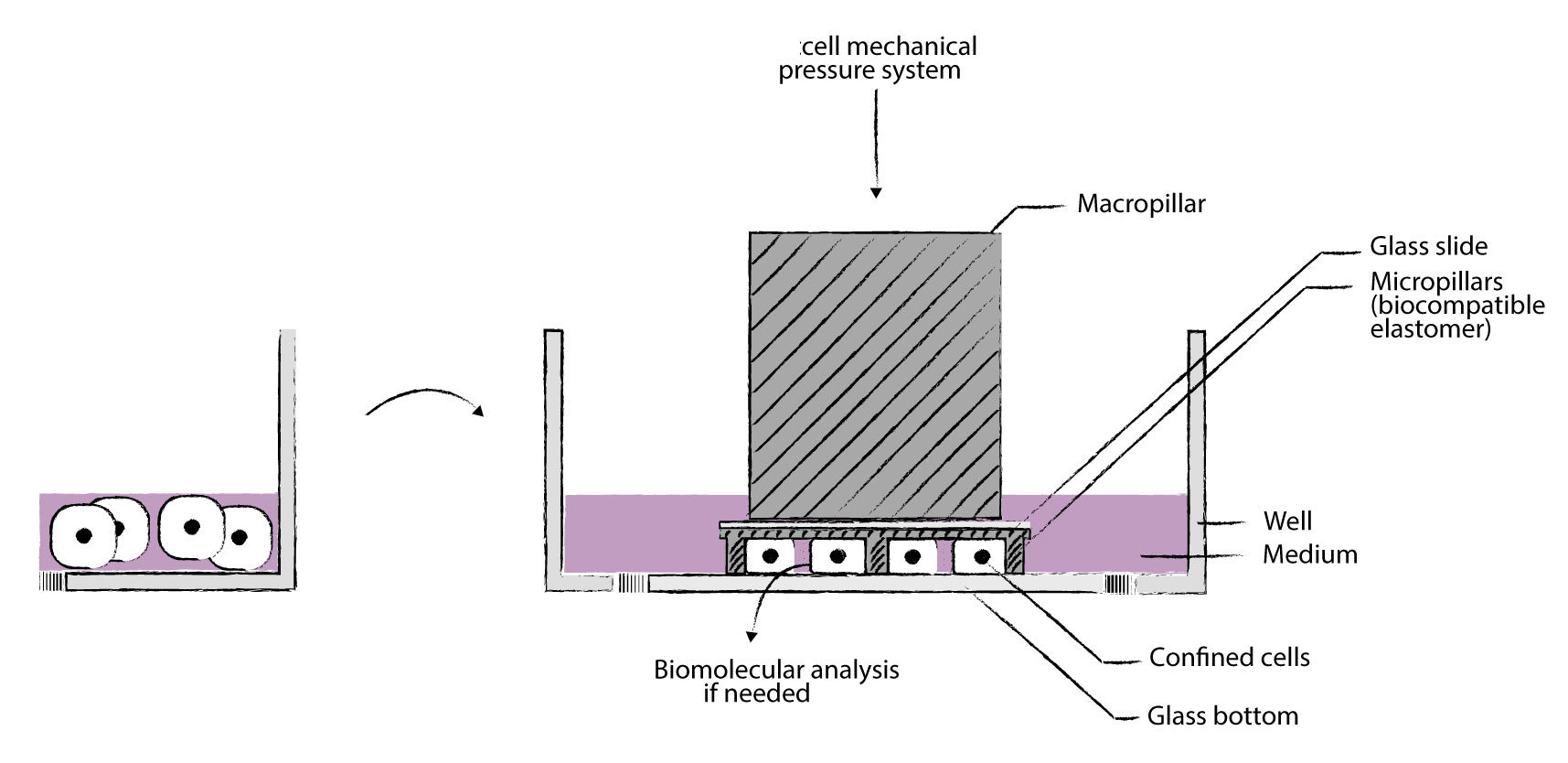
我们的cell Confiner是一种多功能设备,可通过对细胞应用定义明确的约束条件来研究细胞力学。限制方法基于将细胞固定在两个平行表面之间,从而实现均匀且定义明确的物理参数,例如细胞几何形状和环境弹性。此外,可以使用高分辨率显微镜对受限的细胞进行成像,因为该设备是光学透明的,并将细胞保持在焦平面上。
细胞被均匀地限制/压缩在两个亚微米分辨率的两个平行表面之间。两个表面之间的空间由微型PDMS支柱控制。 微型支柱在载玻片上制造,载玻片连接到PDMS活塞(吸盘)上。 活塞由真空泵控制,因此限制区的高度也受到控制。不同的限制高度(例如1um – 300um),允许长期细胞培养和细胞增殖,同时保持对封闭的完美控制与高分辨率光学显微镜系统兼容,可以处理足够多的细胞以进行完整的基因表达分析,可与生物功能化的微结构化底物和/或不同的基质(几何形状控制)结合使用
可以与凝胶结合(硬度控制),兼容任何细胞培养底物(培养皿至96孔板)。
A cell life under confinement背景及重要性
体内细胞被其他细胞和细胞外基质包围并不断受到力的现实。特定细胞会经历其他类型的应变:肌肉、心脏和肺细胞承受各种类型的重复应变,并且肿瘤环境承受增加的压力。
我们的细胞限制器可以解决这个问题。该细胞限制器再现了细胞在体内通常所处的条件:它们被限制。它允许您控制细胞的厚度和细胞培养物的体积。
通过非破坏性方法,您可以精确定义培养物的体积以及施加在细胞上的特定压力。这构成了一个新的相关系统来培养触发仿生的细胞
该细胞限制器是一种能够以微米精度将细胞限制在两个表面内的装置。两个表面之间的空间通过使用微柱来控制。
微柱制造在顶部限制表面(载玻片)中。载玻片连接到充当活塞的 PDMS 结构。
该活塞由真空泵控制,从而也控制了限制的高度。该装置可用于培养皿、玻璃基板等。
特点:
细胞被均匀地限制/压缩亚微米分辨率微米高度两个平行表面之间。
提供不同的限制高度(例如 1 µm – 300um)
允许长期细胞培养和细胞增殖进行,同时保持对限制的完美控制
与高分辨率光学显微镜系统兼容
可以处理足够多的细胞以进行全基因表达分析
可与生物功能化微结构基材和/或不同基质(几何控制)结合
可与凝胶结合(硬度控制)
兼容 35 mm 和 6 孔板的培养皿
技术方案图
三种产品形式
单孔静态细胞限制器
单孔动态细胞限制器
六孔静态细胞限制器
典型应用:
单孔静态细胞限制器
单孔动态细胞限制器
六孔静态细胞限制器
典型应用:
>癌症浸润测定:迁移行为和迁移转变的量化
>癌症侵袭性测定:体细胞或癌细胞的收缩力定量
>内吞作用测定:更好地观察膜发生的事件
>胞吐法测定:更好地观察在顶端膜发生的事件
>吞噬功能失调:机制的表征
>孔中的免疫系统:非粘附免疫细胞的二维迁移和相互作用
>免疫细胞相互作用:非贴壁免疫细胞的2D相互作用
>有丝分裂组装测定:有丝分裂纺锤体疾病的定量
>定量细胞迁移测定:细胞迁移特性的快速,精细分析
>癌症研究转移细胞的迁移
转移中的细胞收缩
DNA DSB修复(机械诱导)
基因组不稳定(细胞分裂)
分离共培养
>免疫学
免疫细胞迁移
非粘附细胞的成像
>器官生理学
癌细胞迁移
具有硬度控制的细胞区分
伤口愈合
分离共培养
细胞压缩反应
>罕见疾病
细胞核完整性
>老化
细胞核完整性
自噬相关疾病
>观测化
非粘附细胞的成像
细胞器的平面成像
>基础研究
细胞体积(细胞周期)
细胞机械力刺激反应
二维心肌细胞成熟测定
二维肝小管化验
3D心肌细胞成熟测定
3D肝小管测定
附着球体测定
细胞收缩力测定
细胞迁移测定
细胞核挤压测定
细胞j化
细胞体积测量
趋化性测定
共培养测定
胞吞试验
胞吐法
外泌体测定
片状脂蛋白和丝状体含量测定
活细胞成像
巨噬细胞j化测定
MT依赖性运输测定
神经肌肉连接测定
井中的神经元网络
细胞器定位分析
初次纤毛测定
骨骼肌细胞测定
平滑肌细胞
伤口愈合测定
-
Confinement and Low Adhesion Induce Fast Amoeboid Migration of Slow Mesenchymal Cells
Y.-J. Liu, M. Piel, Cell, et al., 2015 160(4), 659-672 -
Actin flows induce a universal coupling between cell speed and cell persistence
P. Maiuri, R. Voituriez, et al., Cell, 2015 161(2), 374–386 -
Geometric friction directs cell migration
M. Le Berre, M. Piel, et al., Physical Review Letter 2013 111, 198101 -
Mitotic rounding alters cell geometry to ensure efficient spindle assembly
O. M. Lancaster, B. Baum, et al., Developmental Cell, 2013 25(3), 270-283 -
Fine Control of Nuclear Confinement Identifies a Threshold Deformation leading to Lamina Rupture and Induction of Specific Genes
M. Le Berre, J. Aubertin, M. Piel, Integrative Biology, 2012 4 (11), 1406-1414 -
Exploring the Function of Cell Shape and Size during Mitosis
C. Cadart, H. K. Matthews, et al., Developmental Cell, 2014 29(2), 159-169 -
Methods for Two-Dimensional Cell Confinement
M. Le Berre, M. Piel, et al., 2014, Micropatterning in Cell Biology Part C, Methods in cell biology, 121, 213-29
References
[1] D. Huh, G. A. Hamilton, and D. E. Ingber, “From 3D cell culture to organs-on-chips,” Trends Cell Biol., vol. 21, no. 12, pp. 745–754, 2011.
[2] M. Ravi, V. Paramesh, S. R. Kaviya, E. Anuradha, and F. D. Paul Solomon, “3D cell culture systems: Advantages and applications,” J. Cell. Physiol., vol. 230, no. 1, pp. 16–26, 2015.
[3] J. W. Haycock, 3D cell culture: a review of current approaches and techniques., vol. 695. 2011.
[4] F. Pampaloni, E. G. Reynaud, and E. H. K. Stelzer, “The third dimension bridges the gap between cell culture and live tissue.,” Nat. Rev. Mol. Cell Biol., vol. 8, no. 10, pp. 839–845, 2007.
[5] J. Lee, M. J. Cuddihy, and N. A. Kotov, “Three-dimensional cell culture matrices: state of the art.,” Tissue Eng Part B Rev, vol. 14, no. 1, pp. 61–86, 2008.
[6] M. Vinci et al., “Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation,” BMC Biol., vol. 10, no. 1, p. 29, 2012.
[7] B. A. Justice, N. A. Badr, and R. A. Felder, “3D cell culture opens new dimensions in cell-based assays,” Drug Discov. Today, vol. 14, no. 1–2, pp. 102–107, 2009.
[8] I. Meyvantsson and D. J. Beebe, “Cell culture models in microfluidic systems.,” Annu. Rev. Anal. Chem., vol. 1, pp. 423–449, 2008.
[9] E. W. K. Young and D. J. Beebe, “Fundamentals of microfluidic cell culture in controlled microenvironments,” Chem Soc Rev, vol. 39, no. 3, pp. 1036–1048, 2010.
[10] D. J. Beebe, G. a Mensing, and G. M. Walker, “Physics and applications of microfluidics in biology.,” Annu. Rev. Biomed. Eng., vol. 4, pp. 261–286, 2002.
[11] J. El-Ali, P. K. Sorger, and K. F. Jensen, “Cells on chips.,” Nature, vol. 442, no. 7101, pp. 403–411, 2006.
[12] J. Guck et al., “Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence,” Biophys J, vol. 88, no. 5, pp. 3689–3698, 2005.
[13] S. K?ster et al., “Drop-based microfluidic devices for encapsulation of single cells.,” Lab Chip, vol. 8, no. 7, pp. 1110–1115, 2008.
[14] H. Andersson and A. Van den Berg, “Microfluidic devices for cellomics: A review,” Sensors Actuators, B Chem., vol. 92, no. 3, pp. 315–325, 2003.
[15] M. W. Tibbitt and K. S. Anseth, “Hydrogels as extracellular matrix mimics for 3D cell culture,” Biotechnol. Bioeng., vol. 103, no. 4, pp. 655–663, 2009.
[16] J. P. Vacanti and R. Langer, “Tissue engineering: the design and fabrication of living replacement devices for surgical reconstruction and transplantation.,” Lancet, vol. 354, p. SI32-I34, 1999.
[17] G. S. D. Hetal Patel, Minal Bonde, “Biodegradable polymer scaffolds for tissue engineering,” Trends Biomater. Artif. Organs, vol. 25, no. 1, pp. 20–29, 2011.
[18] L. G. Griffith and M. A. Swartz, “Capturing complex 3D tissue physiology in vitro.,” Nat. Rev. Mol. cell Biol., vol. 7, no. 3, pp. 211–24, 2006.
[19] D. J. Tobin, “Scaffolds for Tissue Engineering and 3D Cell Culture,” Methods Mol. Biol., vol. 695, no. 2, pp. 213–227, 2011.
[20] J. Naranda et al., “Polyester type polyHIPE scaffolds with an interconnected porous structure for cartilage regeneration,” Sci. Rep., vol. 6, no. February, p. 28695, 2016.
[21] B. Dhandayuthapani, Y. Yoshida, T. Maekawa, and D. S. Kumar, “Polymeric scaffolds in tissue engineering application: A review,” Int. J. Polym. Sci., vol. 2011, no. ii, 2011.
[22] F. J. O’Brien, “Biomaterials & scaffolds for tissue engineering,” Mater. Today, vol. 14, no. 3, pp. 88–95, 2011.
[23] A. L. Paguirigan and D. J. Beebe, “Microfluidics meet cell biology: Bridging the gap by validation and application of microscale techniques for cell biological assays,” BioEssays, vol. 30, no. 9, pp. 811–821, Sep. 2008.
[24] F.-Q. Nie, M. Yamada, J. Kobayashi, M. Yamato, A. Kikuchi, and T. Okano, “On-chip cell migration assay using microfluidic channels.,” Biomaterials, vol. 28, no. 27, pp. 4017–4022, 2007.
[25] A. Valster, N. L. Tran, M. Nakada, M. E. Berens, A. Y. Chan, and M. Symons, “Cell migration and invasion assays,” Methods, vol. 37, no. 2, pp. 208–215, 2005.
[26] C. R. Justus, N. Leffler, M. Ruiz-Echevarria, and L. V Yang, “In vitro cell migration and invasion assays.,” J. Vis. Exp., vol. 752, no. 88, p. e51046, 2014.
[27] N. Kramer et al., “In vitro cell migration and invasion assays.,” Mutat Res, vol. 752, no. 1, pp. 10–24, 2013.
[28] J. W. Hong, V. Studer, G. Hang, W. F. Anderson, and S. R. Quake, “A nanoliter-scale nucleic acid processor with parallel architecture.,” Nat. Biotechnol., vol. 22, no. 4, pp. 435–439, 2004.
[29] J. Q. Boedicker, L. Li, T. R. Kline, and R. F. Ismagilov, “Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics.,” Lab Chip, vol. 8, no. 8, pp. 1265–1272, 2008.
[30] G. Velve-Casquillas, M. Le Berre, M. Piel, and P. T. Tran, “Microfluidic tools for cell biological research,” Nano Today, vol. 5, no. 1. pp. 28–47, 2010.
[31] C. R. Terenna et al., “Physical Mechanisms Redirecting Cell Polarity and Cell Shape in Fission Yeast,” Curr. Biol., vol. 18, no. 22, pp. 1748–1753, Nov. 2008.
[32] G. Faure-andré, “Regulation of Dendritic Cell Migration by CD74, the MHC Class II–Associated Invariant Chain,” Science (80-. )., vol. 1705, no. December, 2008.
[33] S. M. McFaul, B. K. Lin, and H. Ma, “Cell separation based on size and deformability using microfluidic funnel ratchets,” Lab Chip, vol. 12, no. 13, pp. 2369–2376, 2012.
[34] S. C. Hur, N. K. Henderson-MacLennan, E. R. B. McCabe, and D. Di Carlo, “Deformability-based cell classification and enrichment using inertial microfluidics.,” Lab Chip, vol. 11, no. 5, pp. 912–920, 2011.
[35] H. W. Hou, Q. S. Li, G. Y. H. Lee, A. P. Kumar, C. N. Ong, and C. T. Lim, “Deformability study of breast cancer cells using microfluidics,” Biomed. Microdevices, vol. 11, no. 3, pp. 557–564, 2009.